| oligos_5-8nt_m5_shift9 (oligos_5-8nt_m5) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
; oligos_5-8nt_m5; m=0 (reference); ncol1=13; shift=9; ncol=23; ---------mmmcACGCGctyy-
; Alignment reference
a 0 0 0 0 0 0 0 0 0 16 17 37 13 55 2 2 4 3 3 8 11 14 0
c 0 0 0 0 0 0 0 0 0 24 21 18 42 0 59 1 56 1 40 6 17 17 0
g 0 0 0 0 0 0 0 0 0 10 13 5 4 7 1 58 0 57 8 9 13 11 0
t 0 0 0 0 0 0 0 0 0 13 12 3 4 1 1 2 3 2 12 40 22 21 0
|
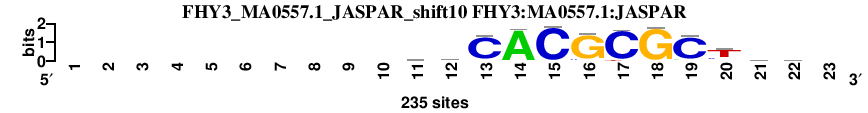
| FHY3_MA0557.1_JASPAR_shift10 (FHY3:MA0557.1:JASPAR) |
 |
0.952 |
0.879 |
5.574 |
0.960 |
0.981 |
1 |
1 |
7 |
1 |
1 |
2.200 |
1 |
; oligos_5-8nt_m5 versus FHY3_MA0557.1_JASPAR (FHY3:MA0557.1:JASPAR); m=1/45; ncol2=12; w=12; offset=1; strand=D; shift=10; score= 2.2; ----------ywCACGCGCThw-
; cor=0.952; Ncor=0.879; logoDP=5.574; NsEucl=0.960; NSW=0.981; rcor=1; rNcor=1; rlogoDP=7; rNsEucl=1; rNSW=1; rank_mean=2.200; match_rank=1
a 0 0 0 0 0 0 0 0 0 0 46 75 9 226 2 3 0 4 18 16 63 82 0
c 0 0 0 0 0 0 0 0 0 0 81 58 210 3 230 16 220 0 208 40 68 44 0
g 0 0 0 0 0 0 0 0 0 0 40 25 9 2 1 213 0 228 6 17 40 50 0
t 0 0 0 0 0 0 0 0 0 0 68 77 7 4 2 3 15 3 3 162 64 59 0
|
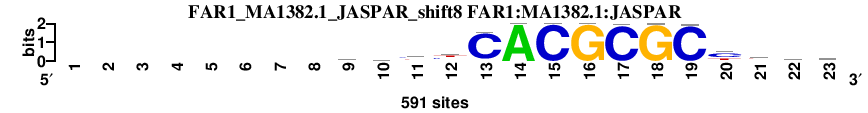
| FAR1_MA1382.1_JASPAR_shift8 (FAR1:MA1382.1:JASPAR) |
 |
0.897 |
0.777 |
6.894 |
0.943 |
0.957 |
5 |
3 |
1 |
5 |
5 |
3.800 |
2 |
; oligos_5-8nt_m5 versus FAR1_MA1382.1_JASPAR (FAR1:MA1382.1:JASPAR); m=2/45; ncol2=15; w=13; offset=-1; strand=D; shift=8; score= 3.8; --------ayyyCACGCGCytyy
; cor=0.897; Ncor=0.777; logoDP=6.894; NsEucl=0.943; NSW=0.957; rcor=5; rNcor=3; rlogoDP=1; rNsEucl=5; rNSW=5; rank_mean=3.800; match_rank=2
a 0 0 0 0 0 0 0 0 234 122 73 126 8 591 0 0 0 0 0 27 112 145 135
c 0 0 0 0 0 0 0 0 117 191 236 193 550 0 591 0 587 0 586 321 62 212 219
g 0 0 0 0 0 0 0 0 98 97 58 13 17 0 0 591 0 590 0 50 147 82 66
t 0 0 0 0 0 0 0 0 142 181 224 259 16 0 0 0 4 1 5 193 270 152 171
|
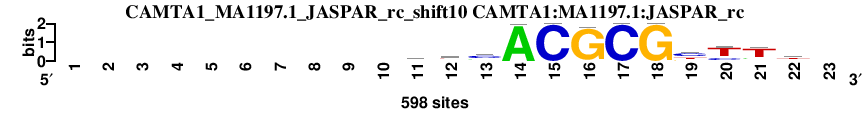
| CAMTA1_MA1197.1_JASPAR_rc_shift10 (CAMTA1:MA1197.1:JASPAR_rc) |
 |
0.926 |
0.855 |
5.302 |
0.952 |
0.972 |
4 |
2 |
10 |
2 |
4 |
4.400 |
3 |
; oligos_5-8nt_m5 versus CAMTA1_MA1197.1_JASPAR_rc (CAMTA1:MA1197.1:JASPAR_rc); m=3/45; ncol2=12; w=12; offset=1; strand=R; shift=10; score= 4.4; ----------yhmACGCGytTt-
; cor=0.926; Ncor=0.855; logoDP=5.302; NsEucl=0.952; NSW=0.972; rcor=4; rNcor=2; rlogoDP=10; rNsEucl=2; rNSW=4; rank_mean=4.400; match_rank=3
a 0 0 0 0 0 0 0 0 0 0 133 174 151 595 0 20 0 0 1 55 77 124 0
c 0 0 0 0 0 0 0 0 0 0 170 156 326 3 598 0 598 0 285 132 45 117 0
g 0 0 0 0 0 0 0 0 0 0 58 33 58 0 0 578 0 598 140 6 46 56 0
t 0 0 0 0 0 0 0 0 0 0 237 235 63 0 0 0 0 0 172 405 430 301 0
|
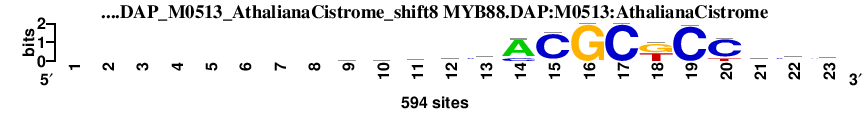
| MYB88.DAP_M0513_AthalianaCistrome_shift8 (MYB88.DAP:M0513:AthalianaCistrome) |
 |
0.792 |
0.687 |
4.741 |
0.930 |
0.936 |
14 |
4 |
15 |
9 |
12 |
10.800 |
5 |
; oligos_5-8nt_m5 versus MYB88.DAP_M0513_AthalianaCistrome (MYB88.DAP:M0513:AthalianaCistrome); m=5/45; ncol2=15; w=13; offset=-1; strand=D; shift=8; score= 10.8; --------hmymcACGCkCCwcy
; cor=0.792; Ncor=0.687; logoDP=4.741; NsEucl=0.930; NSW=0.936; rcor=14; rNcor=4; rlogoDP=15; rNsEucl=9; rNSW=12; rank_mean=10.800; match_rank=5
a 0 0 0 0 0 0 0 0 177 161 122 214 82 472 21 0 0 0 0 5 160 100 108
c 0 0 0 0 0 0 0 0 151 179 217 174 304 111 545 0 594 0 588 482 61 296 276
g 0 0 0 0 0 0 0 0 101 113 87 59 95 11 28 594 0 301 0 5 122 70 61
t 0 0 0 0 0 0 0 0 165 141 168 147 113 0 0 0 0 293 6 102 251 128 149
|




